我就废话不多说了,大家还是直接看代码吧~
# encoding=utf8 ''' 查看和显示nii文件 ''' import matplotlib matplotlib.use('TkAgg') from matplotlib import pylab as plt import nibabel as nib from nibabel import nifti1 from nibabel.viewers import OrthoSlicer3D example_filename = '../ADNI_nii/ADNI_002_S_0413_MR_MPR____N3__Scaled_2_Br_20081001114937668_S14782_I118675.nii' img = nib.load(example_filename) print (img) print (img.header['db_name']) #输出头信息 width,height,queue=img.dataobj.shape OrthoSlicer3D(img.dataobj).show() num = 1 for i in range(0,queue,10): img_arr = img.dataobj[:,:,i] plt.subplot(5,4,num) plt.imshow(img_arr,cmap='gray') num +=1 plt.show()3D显示结果:
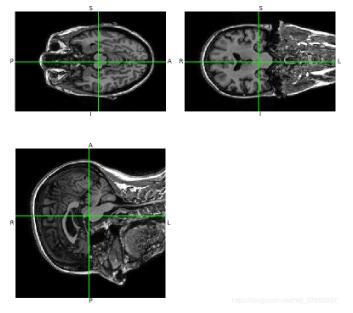
ADNI数据维度(256,256,170)分段显示:
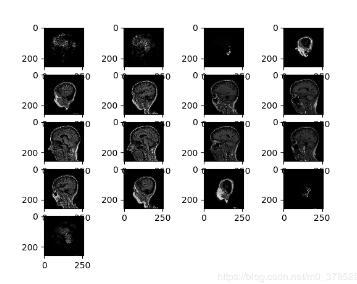
补充知识:python nii图像扩充
我就废话不多说了,大家还是直接看代码吧~
import os import nibabel as nib import numpy as np import math src_us_folder = 'F:/src/ori' src_seg_folder = 'G:/src/seg' aug_us_folder = 'G:/aug/ori' aug_seg_folder = 'G:/aug/seg' img_n= 10 rotate_theta = np.array([0, math.pi/2]) # augmentation aug_cnt = 0 for k in range(img_n): src_us_file = os.path.join(src_us_folder, (str(k) + '.nii')) src_seg_file = os.path.join(src_seg_folder, (str(k) + '_seg.nii')) # load .nii files src_us_vol = nib.load(src_us_file) src_seg_vol = nib.load(src_seg_file) # volume data us_vol_data = src_us_vol.get_data() us_vol_data = (np.array(us_vol_data)).astype('uint8') seg_vol_data = src_seg_vol.get_data() seg_vol_data = (np.array(seg_vol_data)).astype('uint8') # get refer affine matrix ref_affine = src_us_vol.affine ############### flip volume ############### flip_us_vol = np.fliplr(us_vol_data) flip_seg_vol = np.fliplr(seg_vol_data) # construct new volumes new_us_vol = nib.Nifti1Image(flip_us_vol, ref_affine) new_seg_vol = nib.Nifti1Image(flip_seg_vol, ref_affine) # save aug_us_file = os.path.join(aug_us_folder, (str(aug_cnt) + '.nii')) aug_seg_file = os.path.join(aug_seg_folder, (str(aug_cnt) + '_seg.nii')) nib.save(new_us_vol, aug_us_file) nib.save(new_seg_vol, aug_seg_file) aug_cnt = aug_cnt + 1 ############### rotate volume ############### for t in range(len(rotate_theta)): print 'rotating %d theta of %d volume...' % (t, k) cos_gamma = np.cos(t) sin_gamma = np.sin(t) rot_affine = np.array([[1, 0, 0, 0], [0, cos_gamma, -sin_gamma, 0], [0, sin_gamma, cos_gamma, 0], [0, 0, 0, 1]]) new_affine = rot_affine.dot(ref_affine) # construct new volumes new_us_vol = nib.Nifti1Image(us_vol_data, new_affine) new_seg_vol = nib.Nifti1Image(seg_vol_data, new_affine) # save aug_us_file = os.path.join(aug_us_folder, (str(aug_cnt) + '.nii')) aug_seg_file = os.path.join(aug_seg_folder, (str(aug_cnt) + '_seg.nii')) nib.save(new_us_vol, aug_us_file) nib.save(new_seg_vol, aug_seg_file) aug_cnt = aug_cnt + 1以上这篇python 读取.nii格式图像实例就是小编分享给大家的全部内容了,希望能给大家一个参考,也希望大家多多支持python博客。
-
<< 上一篇 下一篇 >>
标签:numpy matplotlib
python 读取.nii格式图像实例
看: 1476次 时间:2020-09-03 分类 : python教程
- 相关文章
- 2021-12-20Python 实现图片色彩转换案例
- 2021-12-20python初学定义函数
- 2021-12-20图文详解Python如何导入自己编写的py文件
- 2021-12-20python二分法查找实例代码
- 2021-12-20Pyinstaller打包工具的使用以及避坑
- 2021-12-20Facebook开源一站式服务python时序利器Kats详解
- 2021-12-20pyCaret效率倍增开源低代码的python机器学习工具
- 2021-12-20python机器学习使数据更鲜活的可视化工具Pandas_Alive
- 2021-12-20python读写文件with open的介绍
- 2021-12-20Python生成任意波形并存为txt的实现
-
搜索
-
-
推荐资源
-
Powered By python教程网 鲁ICP备18013710号
python博客 - 小白学python最友好的网站!